This templated class represents a Statistical Inverse Problem. More...
#include <StatisticalInverseProblem.h>
Public Member Functions | |
Constructor/Destructor methods | |
| StatisticalInverseProblem (const char *prefix, const SipOptionsValues *alternativeOptionsValues, const BaseVectorRV< P_V, P_M > &priorRv, const BaseScalarFunction< P_V, P_M > &likelihoodFunction, GenericVectorRV< P_V, P_M > &postRv) | |
| Constructor. More... | |
| StatisticalInverseProblem (const char *prefix, const SipOptionsValues *alternativeOptionsValues, const GPMSAFactory< P_V, P_M > &gpmsaFactory, GenericVectorRV< P_V, P_M > &postRv) | |
| Constructor for statistical inverse problems to be sovled using GPMSA. More... | |
| ~StatisticalInverseProblem () | |
| Destructor. More... | |
Statistical methods | |
| bool | computeSolutionFlag () const |
| Whether or not compute the solution. More... | |
| void | solveWithBayesMetropolisHastings (const MhOptionsValues *alternativeOptionsValues, const P_V &initialValues, const P_M *initialProposalCovMatrix) |
| Solves the problem via Bayes formula and a Metropolis-Hastings algorithm. More... | |
| void | seedWithMAPEstimator () |
| Seeds the chain with the result of a deterministic optimisation. More... | |
| void | solveWithBayesMLSampling () |
| Solves with Bayes Multi-Level (ML) sampling. More... | |
| const MetropolisHastingsSG < P_V, P_M > & | sequenceGenerator () const |
| Return the underlying MetropolisHastingSG object. More... | |
| const BaseVectorRV< P_V, P_M > & | priorRv () const |
| Returns the Prior RV; access to private attribute m_priorRv. More... | |
| const GenericVectorRV< P_V, P_M > & | postRv () const |
| Returns the Posterior RV; access to private attribute m_postrRv. More... | |
| const BaseVectorSequence< P_V, P_M > & | chain () const |
| Returns the MCMC chain; access to private attribute m_chain. More... | |
| const ScalarSequence< double > & | logLikelihoodValues () const |
| Returns log likelihood values; access to private attribute m_logLikelihoodValues. More... | |
| const ScalarSequence< double > & | logTargetValues () const |
| Returns log target values; access to private attribute m_logTargetValues. More... | |
| double | logEvidence () const |
| Returns the logarithm value of the evidence. Related to ML. More... | |
| double | meanLogLikelihood () const |
| Returns the mean of the logarithm value of the likelihood. Related to ML. More... | |
| double | eig () const |
Private Attributes | |
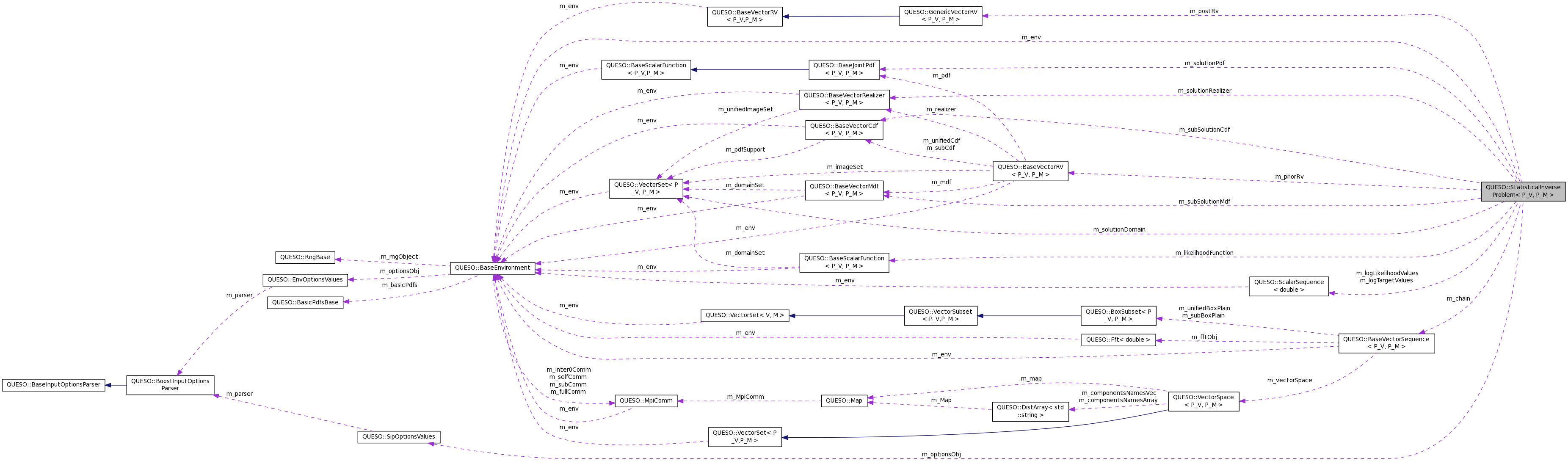
| const BaseEnvironment & | m_env |
| const BaseVectorRV< P_V, P_M > & | m_priorRv |
| const BaseScalarFunction< P_V, P_M > & | m_likelihoodFunction |
| GenericVectorRV< P_V, P_M > & | m_postRv |
| VectorSet< P_V, P_M > * | m_solutionDomain |
| BaseJointPdf< P_V, P_M > * | m_solutionPdf |
| BaseVectorMdf< P_V, P_M > * | m_subSolutionMdf |
| BaseVectorCdf< P_V, P_M > * | m_subSolutionCdf |
| BaseVectorRealizer< P_V, P_M > * | m_solutionRealizer |
| MetropolisHastingsSG< P_V, P_M > * | m_mhSeqGenerator |
| MLSampling< P_V, P_M > * | m_mlSampler |
| BaseVectorSequence< P_V, P_M > * | m_chain |
| ScalarSequence< double > * | m_logLikelihoodValues |
| ScalarSequence< double > * | m_logTargetValues |
| const SipOptionsValues * | m_optionsObj |
| bool | m_seedWithMAPEstimator |
| bool | m_userDidNotProvideOptions |
I/O methods | |
| void | print (std::ostream &os) const |
| TODO: Prints the sequence. More... | |
| std::ostream & | operator<< (std::ostream &os, const StatisticalInverseProblem< P_V, P_M > &obj) |
Detailed Description
template<class P_V = GslVector, class P_M = GslMatrix>
class QUESO::StatisticalInverseProblem< P_V, P_M >
This templated class represents a Statistical Inverse Problem.
This class is templated on the type 'P_V' of vector and type 'P_M' of matrix, where 'P_' stands for 'parameter'. Some templated classes might also use 'Q_V' and 'Q_M' when referring to a vector type and a matrix type, where 'Q_' stands for 'quantities of interest'. Conceptually, a statistical inverse problem has two input entities and one output entity.
The input entities of a statistical inverse problem are:
- the prior RV, an instance of class 'BaseVectorRV<P_V,P_M>', and
- the likelihood function, an instance of class 'BaseScalarFunction<P_V,P_M>'.
Let 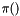 denote the mathematical likelihood function and
denote the mathematical likelihood function and 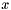 denote a vector of parameters. The likelihood function object stores the routine that computes
denote a vector of parameters. The likelihood function object stores the routine that computes 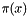 and whatever data necessary by such routine. The routine in the likelihood function object can compute either the actual value
and whatever data necessary by such routine. The routine in the likelihood function object can compute either the actual value 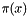 or the value
or the value ![$ \ln[\pi(x)] $](form_157.png) . See files 'basic/inc/ScalarFunction.h' and 'stats/inc/JointPdf.h' for more details.
. See files 'basic/inc/ScalarFunction.h' and 'stats/inc/JointPdf.h' for more details.
The output entity of a statistical inverse problem is the posterior RV, another instance of class 'BaseVectorRV<P_V,P_M>', which stores the solution according to the Bayesian approach. Upon return from a solution operation, the posterior RV is available through the operation 'postRv()'. Such posterior RV is able to provide:
- a joint pdf (up to a multiplicative constant) through the operation 'postRv().pdf()', which returns an instance of the class 'BaseJointPdf<P_V,P_M>', and
- a vector realizer through the operation 'postRv().realizer()', which returns an instance of the class 'BaseVectorRealizer<P_V,P_M>'.
Definition at line 84 of file StatisticalInverseProblem.h.
Constructor & Destructor Documentation
| QUESO::StatisticalInverseProblem< P_V, P_M >::StatisticalInverseProblem | ( | const char * | prefix, |
| const SipOptionsValues * | alternativeOptionsValues, | ||
| const BaseVectorRV< P_V, P_M > & | priorRv, | ||
| const BaseScalarFunction< P_V, P_M > & | likelihoodFunction, | ||
| GenericVectorRV< P_V, P_M > & | postRv | ||
| ) |
Constructor.
Requirements: 1) the image set of the vector random variable 'priorRv', 2) the domain set of the likelihood function 'likelihoodFunction' and 3) the image set of the vector random variable 'postRv' should belong to vector spaces of equal dimensions. If the requirements are satisfied, the constructor then reads input options that begin with the string '<prefix>ip_'. If no options input file is provided, the construction assigns alternativeOptionsValues to the options of the SIP.
- Parameters
-
prefix The prefix alternativeOptionsValues Options (if no input file) priorRv The prior RV likelihoodFunction The likelihood function postRv The posterior RV
Definition at line 35 of file StatisticalInverseProblem.C.
References queso_require_equal_to_msg.
| QUESO::StatisticalInverseProblem< P_V, P_M >::StatisticalInverseProblem | ( | const char * | prefix, |
| const SipOptionsValues * | alternativeOptionsValues, | ||
| const GPMSAFactory< P_V, P_M > & | gpmsaFactory, | ||
| GenericVectorRV< P_V, P_M > & | postRv | ||
| ) |
Constructor for statistical inverse problems to be sovled using GPMSA.
Requirements: 1) the factory for the GPMSA object 2) the image set of the vector random variable postRv (obtainable via GaussianProcessFactory::prior method) should be equal to the full prior image set (including all the hyperparameters)
If the requirements are satisfied, the constructor then reads input options that begin with the string '<prefix>ip_'. If no options input file is provided, the construction assigns alternativeOptionsValues to the options of the statistical inverse problem.
Definition at line 108 of file StatisticalInverseProblem.C.
References QUESO::VectorSpace< V, M >::dimLocal(), QUESO::BaseScalarFunction< V, M >::domainSet(), QUESO::BaseVectorRV< V, M >::imageSet(), QUESO::StatisticalInverseProblem< P_V, P_M >::m_env, QUESO::SipOptionsValues::m_help, QUESO::StatisticalInverseProblem< P_V, P_M >::m_likelihoodFunction, QUESO::StatisticalInverseProblem< P_V, P_M >::m_optionsObj, QUESO::SipOptionsValues::m_prefix, QUESO::StatisticalInverseProblem< P_V, P_M >::m_priorRv, QUESO::StatisticalInverseProblem< P_V, P_M >::m_userDidNotProvideOptions, QUESO::BaseEnvironment::optionsInputFileName(), queso_require_equal_to_msg, QUESO::BaseEnvironment::subDisplayFile(), and QUESO::VectorSet< V, M >::vectorSpace().
| QUESO::StatisticalInverseProblem< P_V, P_M >::~StatisticalInverseProblem | ( | ) |
Destructor.
Definition at line 172 of file StatisticalInverseProblem.C.
Member Function Documentation
| const BaseVectorSequence< P_V, P_M > & QUESO::StatisticalInverseProblem< P_V, P_M >::chain | ( | ) | const |
Returns the MCMC chain; access to private attribute m_chain.
Only valid after solve has been called.
Definition at line 456 of file StatisticalInverseProblem.C.
References queso_require_msg.
| bool QUESO::StatisticalInverseProblem< P_V, P_M >::computeSolutionFlag | ( | ) | const |
Whether or not compute the solution.
| double QUESO::StatisticalInverseProblem< P_V, P_M >::eig | ( | ) | const |
Definition at line 493 of file StatisticalInverseProblem.C.
References queso_require_msg.
| double QUESO::StatisticalInverseProblem< P_V, P_M >::logEvidence | ( | ) | const |
Returns the logarithm value of the evidence. Related to ML.
Definition at line 479 of file StatisticalInverseProblem.C.
References queso_require_msg.
| const ScalarSequence< double > & QUESO::StatisticalInverseProblem< P_V, P_M >::logLikelihoodValues | ( | ) | const |
Returns log likelihood values; access to private attribute m_logLikelihoodValues.
Only valid for MH and only after solve has been called.
Definition at line 464 of file StatisticalInverseProblem.C.
References queso_require_msg.
| const ScalarSequence< double > & QUESO::StatisticalInverseProblem< P_V, P_M >::logTargetValues | ( | ) | const |
Returns log target values; access to private attribute m_logTargetValues.
Only valid for MH and only after solve has been called.
Definition at line 472 of file StatisticalInverseProblem.C.
References queso_require_msg.
| double QUESO::StatisticalInverseProblem< P_V, P_M >::meanLogLikelihood | ( | ) | const |
Returns the mean of the logarithm value of the likelihood. Related to ML.
Definition at line 486 of file StatisticalInverseProblem.C.
References queso_require_msg.
| const GenericVectorRV< P_V, P_M > & QUESO::StatisticalInverseProblem< P_V, P_M >::postRv | ( | ) | const |
Returns the Posterior RV; access to private attribute m_postrRv.
The Posterior RV contains the solution of the Bayes problem.
Definition at line 449 of file StatisticalInverseProblem.C.
| void QUESO::StatisticalInverseProblem< P_V, P_M >::print | ( | std::ostream & | os | ) | const |
TODO: Prints the sequence.
- Todo:
- : implement me!
Definition at line 501 of file StatisticalInverseProblem.C.
| const BaseVectorRV< P_V, P_M > & QUESO::StatisticalInverseProblem< P_V, P_M >::priorRv | ( | ) | const |
Returns the Prior RV; access to private attribute m_priorRv.
Definition at line 442 of file StatisticalInverseProblem.C.
| void QUESO::StatisticalInverseProblem< P_V, P_M >::seedWithMAPEstimator | ( | ) |
Seeds the chain with the result of a deterministic optimisation.
This only works for Metropolis-Hastings right now. Multi-level is not currently supported.
Definition at line 356 of file StatisticalInverseProblem.C.
| const MetropolisHastingsSG< P_V, P_M > & QUESO::StatisticalInverseProblem< P_V, P_M >::sequenceGenerator | ( | ) | const |
Return the underlying MetropolisHastingSG object.
Definition at line 434 of file StatisticalInverseProblem.C.
| void QUESO::StatisticalInverseProblem< P_V, P_M >::solveWithBayesMetropolisHastings | ( | const MhOptionsValues * | alternativeOptionsValues, |
| const P_V & | initialValues, | ||
| const P_M * | initialProposalCovMatrix | ||
| ) |
Solves the problem via Bayes formula and a Metropolis-Hastings algorithm.
Requirements:
- 'initialValues' should have the same number of components as member variable 'm_priorRv'
- if 'initialProposalCovMatrix' is not NULL, it should be square and its size should be equal to the size of 'initialValues'.
If the requirements are satisfied, this methods checks the member flag 'm_computeSolution' (one of the options read from the input file during construction). If the flag is 'false', the operation returns immediately, computing nothing; otherwise, the operation sets the member variable 'm_postRv' accordingly. The operation:
- sets the pdf of 'm_postRv' equal to an instance of BayesianJointPdf<P_V,P_M>,
- instantiates SequenceOfVectors<P_V,P_M> (the chain),
- instantiates MetropolisHastingsSG<P_V,P_M> (the Metropolis-Hastings algorithm),
- populates the chain with the Metropolis-Hastings algorithm, and
- sets the realizer of 'm_postRv' with the contents of the chain.
If seedWithMapEstimator() is called before this method, then initialValues is used as the initial condition for an optimization procedure, the result of which will be used as the seed for the chain.
Definition at line 201 of file StatisticalInverseProblem.C.
References QUESO::InstantiateIntersection(), QUESO::GslOptimizer::minimize(), QUESO::GslOptimizer::minimizer(), queso_require_equal_to_msg, queso_require_msg, QUESO::GslOptimizer::setInitialPoint(), and UQ_SIP_FILENAME_FOR_NO_FILE.
| void QUESO::StatisticalInverseProblem< P_V, P_M >::solveWithBayesMLSampling | ( | ) |
Solves with Bayes Multi-Level (ML) sampling.
Definition at line 363 of file StatisticalInverseProblem.C.
References QUESO::InstantiateIntersection().
Friends And Related Function Documentation
|
friend |
Definition at line 209 of file StatisticalInverseProblem.h.
Member Data Documentation
|
private |
Definition at line 230 of file StatisticalInverseProblem.h.
|
private |
Definition at line 216 of file StatisticalInverseProblem.h.
Referenced by QUESO::StatisticalInverseProblem< P_V, P_M >::StatisticalInverseProblem().
|
private |
Definition at line 219 of file StatisticalInverseProblem.h.
Referenced by QUESO::StatisticalInverseProblem< P_V, P_M >::StatisticalInverseProblem().
|
private |
Definition at line 231 of file StatisticalInverseProblem.h.
|
private |
Definition at line 232 of file StatisticalInverseProblem.h.
|
private |
Definition at line 228 of file StatisticalInverseProblem.h.
|
private |
Definition at line 229 of file StatisticalInverseProblem.h.
|
private |
Definition at line 234 of file StatisticalInverseProblem.h.
Referenced by QUESO::StatisticalInverseProblem< P_V, P_M >::StatisticalInverseProblem().
|
private |
Definition at line 220 of file StatisticalInverseProblem.h.
|
private |
Definition at line 218 of file StatisticalInverseProblem.h.
Referenced by QUESO::StatisticalInverseProblem< P_V, P_M >::StatisticalInverseProblem().
|
private |
Definition at line 236 of file StatisticalInverseProblem.h.
|
private |
Definition at line 222 of file StatisticalInverseProblem.h.
|
private |
Definition at line 223 of file StatisticalInverseProblem.h.
|
private |
Definition at line 226 of file StatisticalInverseProblem.h.
|
private |
Definition at line 225 of file StatisticalInverseProblem.h.
|
private |
Definition at line 224 of file StatisticalInverseProblem.h.
|
private |
Definition at line 243 of file StatisticalInverseProblem.h.
Referenced by QUESO::StatisticalInverseProblem< P_V, P_M >::StatisticalInverseProblem().
The documentation for this class was generated from the following files:
- src/stats/inc/StatisticalInverseProblem.h
- src/stats/src/StatisticalInverseProblem.C