Class for handling vector samples (sequence of vectors). More...
#include <SequenceOfVectors.h>
Public Types | |
Class typedefs | |
| typedef std::vector< const V * > ::const_iterator | seqVectorPositionConstIteratorTypedef |
| typedef std::vector< const V * > ::iterator | seqVectorPositionIteratorTypedef |
Public Member Functions | |
Constructor/Destructor methods | |
| SequenceOfVectors (const VectorSpace< V, M > &vectorSpace, unsigned int subSequenceSize, const std::string &name) | |
| Default constructor. More... | |
| ~SequenceOfVectors () | |
| Destructor. More... | |
Set methods | |
| SequenceOfVectors< V, M > & | operator= (const SequenceOfVectors< V, M > &rhs) |
Copies values from rhs to this. More... | |
Sequence methods | |
| unsigned int | subSequenceSize () const |
| Size of the sub-sequence of vectors. More... | |
| void | resizeSequence (unsigned int newSubSequenceSize) |
| Resizes the sequence. More... | |
| void | resetValues (unsigned int initialPos, unsigned int numPos) |
Resets a total of numPos values of the sequence starting at position initialPos. More... | |
| void | erasePositions (unsigned int initialPos, unsigned int numPos) |
Erases numPos elements of the sequence starting at position initialPos. More... | |
| void | getPositionValues (unsigned int posId, V &vec) const |
Gets the values of the sequence at position posId and stores them at vec. More... | |
| void | setPositionValues (unsigned int posId, const V &vec) |
Set the values in vec at position posId of the sequence. More... | |
| void | subUniformlySampledCdf (const V &numEvaluationPointsVec, ArrayOfOneDGrids< V, M > &cdfGrids, ArrayOfOneDTables< V, M > &cdfValues) const |
| Uniformly samples from the CDF from the sub-sequence. More... | |
| void | unifiedUniformlySampledCdf (const V &numEvaluationPointsVec, ArrayOfOneDGrids< V, M > &unifiedCdfGrids, ArrayOfOneDTables< V, M > &unifiedCdfValues) const |
| Uniformly samples from the CDF from the sub-sequence. More... | |
| void | subMeanExtra (unsigned int initialPos, unsigned int numPos, V &meanVec) const |
Finds the mean value of the sub-sequence, considering numPos positions starting at position initialPos. More... | |
| void | unifiedMeanExtra (unsigned int initialPos, unsigned int numPos, V &unifiedMeanVec) const |
Finds the mean value of the unified sequence, considering numPos positions starting at position initialPos. More... | |
| void | subMedianExtra (unsigned int initialPos, unsigned int numPos, V &medianVec) const |
Finds the median value of the sub-sequence, considering numPos positions starting at position initialPos. More... | |
| void | unifiedMedianExtra (unsigned int initialPos, unsigned int numPos, V &unifiedMedianVec) const |
Finds the median value of the unfed sequence, considering numPos positions starting at position initialPos. More... | |
| void | subSampleVarianceExtra (unsigned int initialPos, unsigned int numPos, const V &meanVec, V &samVec) const |
Finds the sample variance of the sub-sequence, considering numPos positions starting at position initialPos and of mean meanVec. More... | |
| void | unifiedSampleVarianceExtra (unsigned int initialPos, unsigned int numPos, const V &unifiedMeanVec, V &unifiedSamVec) const |
Finds the sample variance of the unified sequence, considering numPos positions starting at position initialPos and of mean meanVec. More... | |
| void | subSampleStd (unsigned int initialPos, unsigned int numPos, const V &meanVec, V &stdVec) const |
Finds the sample standard deviation of the sub-sequence, considering numPos positions starting at position initialPos and of mean meanVec. More... | |
| void | unifiedSampleStd (unsigned int initialPos, unsigned int numPos, const V &unifiedMeanVec, V &unifiedStdVec) const |
Finds the sample standard deviation of the unified sequence, considering numPos positions starting at position initialPos and of mean meanVec. More... | |
| void | subPopulationVariance (unsigned int initialPos, unsigned int numPos, const V &meanVec, V &popVec) const |
Finds the population variance of the sub-sequence, considering numPos positions starting at position initialPos and of mean meanVec. More... | |
| void | unifiedPopulationVariance (unsigned int initialPos, unsigned int numPos, const V &unifiedMeanVec, V &unifiedPopVec) const |
Finds the population variance of the unified sequence, considering numPos positions starting at position initialPos and of mean meanVec. More... | |
| void | autoCovariance (unsigned int initialPos, unsigned int numPos, const V &meanVec, unsigned int lag, V &covVec) const |
| Calculates the autocovariance. More... | |
| void | autoCorrViaDef (unsigned int initialPos, unsigned int numPos, unsigned int lag, V &corrVec) const |
| Calculates the autocorrelation via definition. More... | |
| void | autoCorrViaFft (unsigned int initialPos, unsigned int numPos, const std::vector< unsigned int > &lags, std::vector< V * > &corrVecs) const |
| Calculates the autocorrelation via Fast Fourier transforms (FFT). More... | |
| void | autoCorrViaFft (unsigned int initialPos, unsigned int numPos, unsigned int numSum, V &autoCorrsSumVec) const |
| Calculates the autocorrelation via Fast Fourier transforms (FFT). More... | |
| void | subMinMaxExtra (unsigned int initialPos, unsigned int numPos, V &minVec, V &maxVec) const |
Finds the minimum and the maximum values of the sub-sequence, considering numPos positions starting at position initialPos. More... | |
| void | unifiedMinMaxExtra (unsigned int initialPos, unsigned int numPos, V &unifiedMinVec, V &unifiedMaxVec) const |
Finds the minimum and the maximum values of the unified sequence, considering numPos positions starting at position initialPos. More... | |
| void | subHistogram (unsigned int initialPos, const V &minVec, const V &maxVec, std::vector< V * > ¢ersForAllBins, std::vector< V * > &quanttsForAllBins) const |
| Calculates the histogram of the sub-sequence. More... | |
| void | unifiedHistogram (unsigned int initialPos, const V &unifiedMinVec, const V &unifiedMaxVec, std::vector< V * > &unifiedCentersForAllBins, std::vector< V * > &unifiedQuanttsForAllBins) const |
| Calculates the histogram of the unified sequence. More... | |
| void | subInterQuantileRange (unsigned int initialPos, V &iqrVec) const |
| Returns the interquartile range of the values in the sub-sequence. More... | |
| void | unifiedInterQuantileRange (unsigned int initialPos, V &unifiedIqrVec) const |
| Returns the interquartile range of the values in the unified sequence. More... | |
| void | subScalesForKde (unsigned int initialPos, const V &iqrVec, unsigned int kdeDimension, V &scaleVec) const |
Selects the scales (bandwidth, scaleVec) for the kernel density estimation, considering only the sub-sequence. More... | |
| void | unifiedScalesForKde (unsigned int initialPos, const V &unifiedIqrVec, unsigned int kdeDimension, V &unifiedScaleVec) const |
| Selects the scales (bandwidth) for the kernel density estimation, considering the unified sequence. More... | |
| void | subGaussian1dKde (unsigned int initialPos, const V &scaleVec, const std::vector< V * > &evalParamVecs, std::vector< V * > &densityVecs) const |
| Gaussian kernel for the KDE estimate of the sub-sequence. More... | |
| void | unifiedGaussian1dKde (unsigned int initialPos, const V &unifiedScaleVec, const std::vector< V * > &unifiedEvalParamVecs, std::vector< V * > &unifiedDensityVecs) const |
| Gaussian kernel for the KDE estimate of the unified sequence. More... | |
| void | subWriteContents (unsigned int initialPos, unsigned int numPos, const std::string &fileName, const std::string &fileType, const std::set< unsigned int > &allowedSubEnvIds) const |
| Writes the sub-sequence to a file. More... | |
| void | subWriteContents (unsigned int initialPos, unsigned int numPos, FilePtrSetStruct &filePtrSet, const std::string &fileType) const |
| Writes the sub-sequence to a file. More... | |
| void | subWriteContents (unsigned int initialPos, unsigned int numPos, std::ofstream &ofs, const std::string &fileType) const |
| Writes the sub-sequence to a file. More... | |
| void | unifiedWriteContents (const std::string &fileName, const std::string &fileType) const |
| void | unifiedReadContents (const std::string &fileName, const std::string &fileType, const unsigned int subSequenceSize) |
| Reads the unified sequence from a file. More... | |
| void | select (const std::vector< unsigned int > &idsOfUniquePositions) |
| TODO: It shall select positions in the sequence of vectors. More... | |
| void | filter (unsigned int initialPos, unsigned int spacing) |
| Filters positions in the sequence of vectors. More... | |
| double | estimateConvBrooksGelman (unsigned int initialPos, unsigned int numPos) const |
| Estimates convergence rate using Brooks & Gelman method. More... | |
| void | extractScalarSeq (unsigned int initialPos, unsigned int spacing, unsigned int numPos, unsigned int paramId, ScalarSequence< double > &scalarSeq) const |
| Extracts a sequence of scalars. More... | |
 Public Member Functions inherited from QUESO::BaseVectorSequence< V, M > Public Member Functions inherited from QUESO::BaseVectorSequence< V, M > | |
| BaseVectorSequence (const VectorSpace< V, M > &vectorSpace, unsigned int subSequenceSize, const std::string &name) | |
| Default constructor. More... | |
| virtual | ~BaseVectorSequence () |
| Destructor. More... | |
| unsigned int | unifiedSequenceSize () const |
| Calculates the size of the unified sequence of vectors. More... | |
| unsigned int | vectorSizeLocal () const |
| Local dimension (size) of the vector space. More... | |
| unsigned int | vectorSizeGlobal () const |
| Global dimension (size) of the vector space. More... | |
| const VectorSpace< V, M > & | vectorSpace () const |
| Vector space; access to protected attribute VectorSpace<V,M>& m_vectorSpace. More... | |
| const std::string & | name () const |
Access to protected attribute m_name: name of the sequence of vectors. More... | |
| void | setName (const std::string &newName) |
| Changes the name of the sequence of vectors. More... | |
| void | clear () |
| Reset the values and the size of the sequence of vectors. More... | |
| const V & | subMinPlain () const |
| Finds the minimum value of the sub-sequence. More... | |
| const V & | unifiedMinPlain () const |
| Finds the minimum value of the unified sequence. More... | |
| const V & | subMaxPlain () const |
| Finds the maximum value of the sub-sequence. More... | |
| const V & | unifiedMaxPlain () const |
| Finds the maximum value of the unified sequence. More... | |
| const V & | subMeanPlain () const |
| Finds the mean value of the sub-sequence. More... | |
| const V & | unifiedMeanPlain () const |
| Finds the mean value of the unified sequence. More... | |
| const V & | subMedianPlain () const |
| Finds the median value of the sub-sequence. More... | |
| const V & | unifiedMedianPlain () const |
| Finds the median value of the unified sequence. More... | |
| const V & | subSampleVariancePlain () const |
| Finds the variance of a sample of the sub-sequence. More... | |
| const V & | unifiedSampleVariancePlain () const |
| Finds the variance of a sample of the unified sequence. More... | |
| const BoxSubset< V, M > & | subBoxPlain () const |
| Finds a box subset of the sub-sequence (given by its min and max values calculated via subMinPlain and subMaxPlain). More... | |
| const BoxSubset< V, M > & | unifiedBoxPlain () const |
| Finds a box subset of the unified-sequence (given by the min and max values of the unified sequence calculated via unifiedMinPlain and unifiedMaxPlain). More... | |
| void | deleteStoredVectors () |
| Deletes all the stored vectors. More... | |
| void | append (const BaseVectorSequence< V, M > &src, unsigned int initialPos, unsigned int numPos) |
Appends the vector src to this vector. More... | |
| double | subPositionsOfMaximum (const ScalarSequence< double > &subCorrespondingScalarValues, BaseVectorSequence< V, M > &subPositionsOfMaximum) |
| Finds the positions where the maximum element occurs in the sub-sequence. More... | |
| double | unifiedPositionsOfMaximum (const ScalarSequence< double > &subCorrespondingScalarValues, BaseVectorSequence< V, M > &unifiedPositionsOfMaximum) |
| Finds the positions where the maximum element occurs in the unified sequence. More... | |
| void | setGaussian (const V &meanVec, const V &stdDevVec) |
Sets the values of the sequence as a Gaussian distribution of mean given by meanVec and standard deviation by stdDevVec. More... | |
| void | setUniform (const V &aVec, const V &bVec) |
Sets the values of the sequence as a uniform distribution between the values given by vectors aVec and bVec. More... | |
| void | computeFilterParams (std::ofstream *passedOfs, unsigned int &initialPos, unsigned int &spacing) |
Computes the filtering parameters spacing for the sequence of vectors. More... | |
Private Member Functions | |
| void | copy (const SequenceOfVectors< V, M > &src) |
Copies vector sequence src to this. More... | |
| void | extractRawData (unsigned int initialPos, unsigned int spacing, unsigned int numPos, unsigned int paramId, std::vector< double > &rawData) const |
| Extracts the raw data. More... | |
| void | writeSubMatlabHeader (std::ofstream &ofs, double sequenceSize, double vectorSizeLocal) const |
| Helper function to write matlab-specific header info for vectors. More... | |
| void | writeUnifiedMatlabHeader (std::ofstream &ofs, double sequenceSize, double vectorSizeLocal) const |
| void | writeTxtHeader (std::ofstream &ofs, double sequenceSize, double vectorSizeLocal) const |
| Helper function to write plain txt info for vectors. More... | |
Private Attributes | |
| std::vector< const V * > | m_seq |
| Sequence of vectors. More... | |
Additional Inherited Members | |
 Protected Member Functions inherited from QUESO::BaseVectorSequence< V, M > Protected Member Functions inherited from QUESO::BaseVectorSequence< V, M > | |
| void | copy (const BaseVectorSequence< V, M > &src) |
Copies vector sequence src to this. More... | |
 Protected Attributes inherited from QUESO::BaseVectorSequence< V, M > Protected Attributes inherited from QUESO::BaseVectorSequence< V, M > | |
| const BaseEnvironment & | m_env |
| const VectorSpace< V, M > & | m_vectorSpace |
| std::string | m_name |
| Fft< double > * | m_fftObj |
| V * | m_subMinPlain |
| V * | m_unifiedMinPlain |
| V * | m_subMaxPlain |
| V * | m_unifiedMaxPlain |
| V * | m_subMeanPlain |
| V * | m_unifiedMeanPlain |
| V * | m_subMedianPlain |
| V * | m_unifiedMedianPlain |
| V * | m_subSampleVariancePlain |
| V * | m_unifiedSampleVariancePlain |
| BoxSubset< V, M > * | m_subBoxPlain |
| BoxSubset< V, M > * | m_unifiedBoxPlain |
Detailed Description
template<class V = GslVector, class M = GslMatrix>
class QUESO::SequenceOfVectors< V, M >
Class for handling vector samples (sequence of vectors).
This class handles vector samples generated by an algorithm, as well as operations that can be carried over them, e.g., calculation of means, correlation and covariance matrices. It is derived from and implements BaseVectorSequence<V,M>.
Definition at line 49 of file SequenceOfVectors.h.
Member Typedef Documentation
| typedef std::vector<const V*>::const_iterator QUESO::SequenceOfVectors< V, M >::seqVectorPositionConstIteratorTypedef |
Definition at line 55 of file SequenceOfVectors.h.
| typedef std::vector<const V*>::iterator QUESO::SequenceOfVectors< V, M >::seqVectorPositionIteratorTypedef |
Definition at line 56 of file SequenceOfVectors.h.
Constructor & Destructor Documentation
| QUESO::SequenceOfVectors< V, M >::SequenceOfVectors | ( | const VectorSpace< V, M > & | vectorSpace, |
| unsigned int | subSequenceSize, | ||
| const std::string & | name | ||
| ) |
Default constructor.
Definition at line 33 of file SequenceOfVectors.C.
| QUESO::SequenceOfVectors< V, M >::~SequenceOfVectors | ( | ) |
Destructor.
Definition at line 68 of file SequenceOfVectors.C.
Member Function Documentation
|
virtual |
Calculates the autocorrelation via definition.
Autocorrelation is the cross-correlation of a variable with itself; it describes the correlation between values of the process at different times, as a function of the two times. It is calculated over a sequence of vectors with initial position initialPos, considering numPos positions, a lag of lag, with mean given by meanVec. Output:
- Parameters
-
corrVec is the vector of the calculated autocorrelations of the sequence of vectors.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 766 of file SequenceOfVectors.C.
References QUESO::ScalarSequence< T >::autoCorrViaDef(), and queso_require_msg.
|
virtual |
Calculates the autocorrelation via Fast Fourier transforms (FFT).
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 798 of file SequenceOfVectors.C.
References QUESO::ScalarSequence< T >::autoCorrViaFft(), and queso_require_msg.
|
virtual |
Calculates the autocorrelation via Fast Fourier transforms (FFT).
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 857 of file SequenceOfVectors.C.
References QUESO::ScalarSequence< T >::autoCorrViaFft(), and queso_require_msg.
|
virtual |
Calculates the autocovariance.
The autocovariance is the covariance of a variable with itself at some other time. It is calculated over a sequence of vectors with initial position initialPos, considering numPos positions, a lag of lag, with mean given by meanVec. Output:
- Parameters
-
covVec is the vector of the calculated autocovariances of the sequence of vectors.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 731 of file SequenceOfVectors.C.
References QUESO::ScalarSequence< T >::autoCovariance(), and queso_require_msg.
|
private |
Copies vector sequence src to this.
Definition at line 2234 of file SequenceOfVectors.C.
References QUESO::BaseVectorSequence< V, M >::copy(), QUESO::SequenceOfVectors< V, M >::m_seq, and QUESO::SequenceOfVectors< V, M >::subSequenceSize().
|
virtual |
Erases numPos elements of the sequence starting at position initialPos.
This routine deletes all stored computed vectors
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 154 of file SequenceOfVectors.C.
References QUESO::BaseVectorSequence< V, M >::deleteStoredVectors(), queso_require_equal_to_msg, and queso_require_msg.
|
virtual |
Estimates convergence rate using Brooks & Gelman method.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 2083 of file SequenceOfVectors.C.
References QUESO::matrixProduct(), queso_require_greater_equal_msg, and work.
|
privatevirtual |
Extracts the raw data.
This method saves in rawData the data from the sequence of vectors (in private attribute m_seq) starting at position (initialPos,paramId) , with a spacing of spacing until numPos positions have been extracted.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 2253 of file SequenceOfVectors.C.
|
virtual |
Extracts a sequence of scalars.
The sequence of scalars has size numPos, and it will be extracted starting at position (initialPos, paramId ) of this sequences of vectors, given spacing spacing.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 2210 of file SequenceOfVectors.C.
References QUESO::ScalarSequence< T >::resizeSequence().
|
virtual |
Filters positions in the sequence of vectors.
Filtered positions will starting at initialPos, and with spacing given by spacing.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 2042 of file SequenceOfVectors.C.
Referenced by QUESO::MLSampling< P_V, P_M >::generateSequence(), and QUESO::MLSampling< P_V, P_M >::generateSequence_Step11_inter0().
|
virtual |
Gets the values of the sequence at position posId and stores them at vec.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 188 of file SequenceOfVectors.C.
References queso_require_less_msg, and queso_require_msg.
Referenced by QUESO::MLSampling< P_V, P_M >::generateBalLinkedChains_all(), QUESO::MLSampling< P_V, P_M >::generateSequence(), QUESO::MetropolisHastingsSG< P_V, P_M >::generateSequence(), QUESO::MLSampling< P_V, P_M >::generateSequence_Step02_inter0(), QUESO::MLSampling< P_V, P_M >::generateSequence_Step04_inter0(), QUESO::MLSampling< P_V, P_M >::generateUnbLinkedChains_all(), QUESO::MLSampling< P_V, P_M >::mpiExchangePositions_inter0(), and QUESO::SimulationModel< S_V, S_M, P_V, P_M, Q_V, Q_M >::SimulationModel().
| SequenceOfVectors< V, M > & QUESO::SequenceOfVectors< V, M >::operator= | ( | const SequenceOfVectors< V, M > & | rhs | ) |
Copies values from rhs to this.
Definition at line 94 of file SequenceOfVectors.C.
References copy.
|
virtual |
Resets a total of numPos values of the sequence starting at position initialPos.
This routine deletes all stored computed vectors
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 126 of file SequenceOfVectors.C.
References QUESO::BaseVectorSequence< V, M >::deleteStoredVectors(), and queso_require_msg.
|
virtual |
Resizes the sequence.
This routine deletes all stored computed vectors
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 110 of file SequenceOfVectors.C.
References QUESO::BaseVectorSequence< V, M >::deleteStoredVectors().
Referenced by QUESO::MetropolisHastingsSG< P_V, P_M >::adapt(), and QUESO::MLSampling< P_V, P_M >::generateSequence_Level0_all().
|
virtual |
TODO: It shall select positions in the sequence of vectors.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 2033 of file SequenceOfVectors.C.
References queso_error_msg.
|
virtual |
Set the values in vec at position posId of the sequence.
This routine deletes all stored computed vectors
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 206 of file SequenceOfVectors.C.
References QUESO::BaseVectorSequence< V, M >::deleteStoredVectors(), queso_require_equal_to_msg, and queso_require_less_msg.
Referenced by QUESO::MetropolisHastingsSG< P_V, P_M >::adapt(), QUESO::GpmsaComputerModel< S_V, S_M, D_V, D_M, P_V, P_M, Q_V, Q_M >::generatePriorSeq(), QUESO::MLSampling< P_V, P_M >::generateSequence_Level0_all(), QUESO::GpmsaComputerModel< S_V, S_M, D_V, D_M, P_V, P_M, Q_V, Q_M >::predictVUsAtGridPoint(), QUESO::GpmsaComputerModel< S_V, S_M, D_V, D_M, P_V, P_M, Q_V, Q_M >::predictWsAtGridPoint(), and QUESO::SimulationModel< S_V, S_M, P_V, P_M, Q_V, Q_M >::SimulationModel().
|
virtual |
Gaussian kernel for the KDE estimate of the sub-sequence.
Computes a probability density estimate of the sample in this sub-sequence, starting at position initialPos. densityVecs is the vector of density values evaluated at the points in evaluationParamVecs. The estimate is based on Gaussian (normal) kernel function, using a window parameter (scaleVec).
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 1165 of file SequenceOfVectors.C.
References queso_require_msg, and QUESO::ScalarSequence< T >::subGaussian1dKde().
|
virtual |
Calculates the histogram of the sub-sequence.
It requires the specification of the number of bins that the histogram will have, the center of each bin and the initial position where the data will be considered.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 956 of file SequenceOfVectors.C.
References queso_require_msg, and QUESO::ScalarSequence< T >::subHistogram().
|
virtual |
Returns the interquartile range of the values in the sub-sequence.
The IQR is a robust estimate of the spread of the data, since changes in the upper and lower 25% of the data do not affect it. If there are outliers in the data, then the IQR is more representative than the standard deviation as an estimate of the spread of the body of the data. The IQR is less efficient than the standard deviation as an estimate of the spread when the data is all from the normal distribution. (from Matlab)
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 1049 of file SequenceOfVectors.C.
References queso_require_msg, and QUESO::ScalarSequence< T >::subInterQuantileRange().
|
virtual |
Finds the mean value of the sub-sequence, considering numPos positions starting at position initialPos.
Output:
- Parameters
-
meanVec is the vector of the calculated means of the sub-sequence of vectors.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 308 of file SequenceOfVectors.C.
References queso_require_msg, and QUESO::ScalarSequence< T >::subMeanExtra().
|
virtual |
Finds the median value of the sub-sequence, considering numPos positions starting at position initialPos.
Output:
- Parameters
-
medianVec is the vector of the calculated medians of the sub-sequence of vectors.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 422 of file SequenceOfVectors.C.
References queso_require_msg, and QUESO::ScalarSequence< T >::subMedianExtra().
|
virtual |
Finds the minimum and the maximum values of the sub-sequence, considering numPos positions starting at position initialPos.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 892 of file SequenceOfVectors.C.
References queso_require_msg, and QUESO::ScalarSequence< T >::subMinMaxExtra().
|
virtual |
Finds the population variance of the sub-sequence, considering numPos positions starting at position initialPos and of mean meanVec.
Output:
- Parameters
-
popVec is the vector of the calculated population variance of the sub-sequence of vectors. The population variance 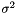 is defined by
is defined by 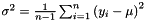 , where
, where 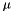 is the sample mean and
is the sample mean and 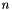 is the sample size. This procedure lets the users choose the initial position and the number of elements of the sequence which will be used to evaluate the population variance .
is the sample size. This procedure lets the users choose the initial position and the number of elements of the sequence which will be used to evaluate the population variance .
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 666 of file SequenceOfVectors.C.
References queso_require_msg, and QUESO::ScalarSequence< T >::subPopulationVariance().
| void QUESO::SequenceOfVectors< V, M >::subSampleStd | ( | unsigned int | initialPos, |
| unsigned int | numPos, | ||
| const V & | meanVec, | ||
| V & | stdVec | ||
| ) | const |
Finds the sample standard deviation of the sub-sequence, considering numPos positions starting at position initialPos and of mean meanVec.
Output:
- Parameters
-
stdVec is the vector of the calculated sample standard deviation of the sub-sequence of vectors.
Definition at line 601 of file SequenceOfVectors.C.
References queso_require_msg, and QUESO::ScalarSequence< T >::subSampleStd().
Referenced by QUESO::SimulationModel< S_V, S_M, P_V, P_M, Q_V, Q_M >::SimulationModel().
|
virtual |
Finds the sample variance of the sub-sequence, considering numPos positions starting at position initialPos and of mean meanVec.
Output:
- Parameters
-
samVec is the vector of the calculated sample variance of the sub-sequence of vectors. The sample variance 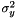 is the second sample central moment and is defined by
is the second sample central moment and is defined by 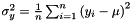 , where
, where 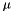 is the sample mean and
is the sample mean and 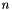 is the sample size. This procedure lets the users choose the initial position and the number of elements of the sequence which will be used to evaluate the sample variance.
is the sample size. This procedure lets the users choose the initial position and the number of elements of the sequence which will be used to evaluate the sample variance.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 536 of file SequenceOfVectors.C.
References queso_require_msg, and QUESO::ScalarSequence< T >::subSampleVarianceExtra().
|
virtual |
Selects the scales (bandwidth, scaleVec) for the kernel density estimation, considering only the sub-sequence.
The bandwidth of the kernel is a free parameter which exhibits a strong influence on the resulting estimate. Silverman (1986) suggests the following normal-based estimates: S1 = 1.06 × (standard deviation) × n^{-1/5} S2 = 0.79 × (iqrVec) × n^{-1/5}, where iqrVec is the interquartile range scaleVec = 0.90 × minimum(standard deviation, iqrVec /1.34) × n^{-1/5}. These estimates are popular due to their simplicity, and are used in QUESO with the adaptation of the exponent oven the sample size n (-1/5) with -1/(4 + kdeDimension) where kdeDimension is the KDE dimension.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 1102 of file SequenceOfVectors.C.
References queso_require_msg, and QUESO::ScalarSequence< T >::subScaleForKde().
|
virtual |
Size of the sub-sequence of vectors.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 103 of file SequenceOfVectors.C.
Referenced by QUESO::MetropolisHastingsSG< P_V, P_M >::adapt(), QUESO::SequenceOfVectors< V, M >::copy(), QUESO::MLSampling< P_V, P_M >::generateBalLinkedChains_all(), QUESO::MLSampling< P_V, P_M >::generateSequence(), QUESO::MetropolisHastingsSG< P_V, P_M >::generateSequence(), QUESO::MLSampling< P_V, P_M >::generateSequence_Level0_all(), QUESO::MLSampling< P_V, P_M >::generateSequence_Step02_inter0(), QUESO::MLSampling< P_V, P_M >::generateSequence_Step11_inter0(), QUESO::MLSampling< P_V, P_M >::generateUnbLinkedChains_all(), QUESO::GpmsaComputerModel< S_V, S_M, D_V, D_M, P_V, P_M, Q_V, Q_M >::predictVUsAtGridPoint(), QUESO::GpmsaComputerModel< S_V, S_M, D_V, D_M, P_V, P_M, Q_V, Q_M >::predictWsAtGridPoint(), and QUESO::SimulationModel< S_V, S_M, P_V, P_M, Q_V, Q_M >::SimulationModel().
| void QUESO::SequenceOfVectors< V, M >::subUniformlySampledCdf | ( | const V & | numEvaluationPointsVec, |
| ArrayOfOneDGrids< V, M > & | cdfGrids, | ||
| ArrayOfOneDTables< V, M > & | cdfValues | ||
| ) | const |
Uniformly samples from the CDF from the sub-sequence.
Definition at line 227 of file SequenceOfVectors.C.
References QUESO::ArrayOfOneDTables< V, M >::setOneDTable(), and QUESO::ArrayOfOneDGrids< V, M >::setUniformGrids().
Referenced by QUESO::StatisticalForwardProblem< P_V, P_M, Q_V, Q_M >::solveWithMonteCarlo().
|
virtual |
Writes the sub-sequence to a file.
Given the allowed sub environments (allowedSubEnvIds) that are allowed to write to file, together with the file name and type (fileName, fileType), it writes the entire sub- sequence to the file. The sum of the initial position of the sequence (initialPos) with the number of positions that will be written (numPos) must equal the size of the sequence. This procedure calls void subWriteContents (unsigned int initialPos, unsigned int numPos, FilePtrSetStruct& filePtrSet, const std::string& fileType).
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 1264 of file SequenceOfVectors.C.
References queso_require_greater_equal_msg.
| void QUESO::SequenceOfVectors< V, M >::subWriteContents | ( | unsigned int | initialPos, |
| unsigned int | numPos, | ||
| FilePtrSetStruct & | filePtrSet, | ||
| const std::string & | fileType | ||
| ) | const |
Writes the sub-sequence to a file.
Uses additional variable of the type FilePtrSetStruct& to operate on files.
Definition at line 1312 of file SequenceOfVectors.C.
References QUESO::FilePtrSetStruct::ofsVar, queso_error_msg, queso_require_greater_equal_msg, queso_require_less_equal_msg, queso_require_msg, UQ_FILE_EXTENSION_FOR_HDF_FORMAT, UQ_FILE_EXTENSION_FOR_MATLAB_FORMAT, and UQ_FILE_EXTENSION_FOR_TXT_FORMAT.
|
virtual |
Writes the sub-sequence to a file.
Uses object of the type std::ofstream.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 1415 of file SequenceOfVectors.C.
References queso_require_less_equal_msg, UQ_FILE_EXTENSION_FOR_MATLAB_FORMAT, and UQ_FILE_EXTENSION_FOR_TXT_FORMAT.
|
virtual |
Gaussian kernel for the KDE estimate of the unified sequence.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 1214 of file SequenceOfVectors.C.
References queso_require_msg, and QUESO::ScalarSequence< T >::unifiedGaussian1dKde().
|
virtual |
Calculates the histogram of the unified sequence.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 1002 of file SequenceOfVectors.C.
References queso_require_msg, and QUESO::ScalarSequence< T >::unifiedHistogram().
|
virtual |
Returns the interquartile range of the values in the unified sequence.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 1075 of file SequenceOfVectors.C.
References queso_require_msg, and QUESO::ScalarSequence< T >::unifiedInterQuantileRange().
|
virtual |
Finds the mean value of the unified sequence, considering numPos positions starting at position initialPos.
Output:
- Parameters
-
meanVec is the vector of the calculated means of the unified sequence of vectors.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 363 of file SequenceOfVectors.C.
References queso_require_msg, and QUESO::ScalarSequence< T >::unifiedMeanExtra().
|
virtual |
Finds the median value of the unfed sequence, considering numPos positions starting at position initialPos.
Output:
- Parameters
-
medianVec is the vector of the calculated medians of the unified sequence of vectors.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 477 of file SequenceOfVectors.C.
References queso_require_msg, and QUESO::ScalarSequence< T >::unifiedMedianExtra().
|
virtual |
Finds the minimum and the maximum values of the unified sequence, considering numPos positions starting at position initialPos.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 922 of file SequenceOfVectors.C.
References queso_require_msg, and QUESO::ScalarSequence< T >::unifiedMinMaxExtra().
|
virtual |
Finds the population variance of the unified sequence, considering numPos positions starting at position initialPos and of mean meanVec.
Output:
- Parameters
-
popVec is the vector of the calculated population variance of the unified sequence of vectors.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 698 of file SequenceOfVectors.C.
References queso_require_msg, and QUESO::ScalarSequence< T >::unifiedPopulationVariance().
|
virtual |
Reads the unified sequence from a file.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 1763 of file SequenceOfVectors.C.
References dataIn, QUESO::FilePtrSetStruct::ifsVar, QUESO::MiscGetEllapsedSeconds(), queso_error_msg, queso_require_equal_to_msg, queso_require_greater_equal_msg, queso_require_less_msg, UQ_FILE_EXTENSION_FOR_HDF_FORMAT, UQ_FILE_EXTENSION_FOR_MATLAB_FORMAT, UQ_FILE_EXTENSION_FOR_TXT_FORMAT, and QUESO::UQ_OK_RC.
Referenced by QUESO::MLSampling< P_V, P_M >::restartML().
| void QUESO::SequenceOfVectors< V, M >::unifiedSampleStd | ( | unsigned int | initialPos, |
| unsigned int | numPos, | ||
| const V & | unifiedMeanVec, | ||
| V & | unifiedStdVec | ||
| ) | const |
Finds the sample standard deviation of the unified sequence, considering numPos positions starting at position initialPos and of mean meanVec.
Output:
- Parameters
-
stdVec is the vector of the calculated sample standard deviation of the unified sequence of vectors.
Definition at line 633 of file SequenceOfVectors.C.
References queso_require_msg, and QUESO::ScalarSequence< T >::unifiedSampleStd().
|
virtual |
Finds the sample variance of the unified sequence, considering numPos positions starting at position initialPos and of mean meanVec.
Output:
- Parameters
-
samVec is the vector of the calculated sample variance of the unified sequence of vectors.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 568 of file SequenceOfVectors.C.
References queso_require_msg, and QUESO::ScalarSequence< T >::unifiedSampleVarianceExtra().
|
virtual |
Selects the scales (bandwidth) for the kernel density estimation, considering the unified sequence.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 1133 of file SequenceOfVectors.C.
References queso_require_msg, and QUESO::ScalarSequence< T >::unifiedScaleForKde().
| void QUESO::SequenceOfVectors< V, M >::unifiedUniformlySampledCdf | ( | const V & | numEvaluationPointsVec, |
| ArrayOfOneDGrids< V, M > & | unifiedCdfGrids, | ||
| ArrayOfOneDTables< V, M > & | unifiedCdfValues | ||
| ) | const |
Uniformly samples from the CDF from the sub-sequence.
Definition at line 262 of file SequenceOfVectors.C.
References QUESO::ArrayOfOneDTables< V, M >::setOneDTable(), and QUESO::ArrayOfOneDGrids< V, M >::setUniformGrids().
Referenced by QUESO::StatisticalForwardProblem< P_V, P_M, Q_V, Q_M >::solveWithMonteCarlo().
|
virtual |
Writes the unfed sequence to a file. Writes the unified sequence in Matlab/Octave format or, if enabled, in HDF5 format.
Implements QUESO::BaseVectorSequence< V, M >.
Definition at line 1493 of file SequenceOfVectors.C.
References QUESO::MiscGetEllapsedSeconds(), QUESO::FilePtrSetStruct::ofsVar, queso_error_msg, UQ_FILE_EXTENSION_FOR_HDF_FORMAT, UQ_FILE_EXTENSION_FOR_MATLAB_FORMAT, UQ_FILE_EXTENSION_FOR_TXT_FORMAT, and QUESO::UQ_OK_RC.
Referenced by QUESO::MLSampling< P_V, P_M >::checkpointML(), QUESO::GpmsaComputerModel< S_V, S_M, D_V, D_M, P_V, P_M, Q_V, Q_M >::generatePriorSeq(), QUESO::MLSampling< P_V, P_M >::generateSequence(), QUESO::MLSampling< P_V, P_M >::generateSequence_Level0_all(), and QUESO::MLSampling< P_V, P_M >::generateSequence_Step11_inter0().
|
private |
Helper function to write matlab-specific header info for vectors.
Definition at line 1459 of file SequenceOfVectors.C.
|
private |
Helper function to write plain txt info for vectors.
Definition at line 1483 of file SequenceOfVectors.C.
|
private |
Definition at line 1471 of file SequenceOfVectors.C.
Member Data Documentation
|
private |
Sequence of vectors.
Definition at line 435 of file SequenceOfVectors.h.
Referenced by QUESO::SequenceOfVectors< V, M >::copy().
The documentation for this class was generated from the following files:
- src/basic/inc/SequenceOfVectors.h
- src/basic/src/SequenceOfVectors.C